Today
Problems with the errors
- Generalized Least Squares
- Lack of fit F-tests
- Robust regression
Generalized Least Squares
We have assumed , but what if we know , where is unknown, but is known. For example, we know the form of the correlation and/or non-constant variance in the response.
The usual least squares estimates are unbiased, but they are no longer BLUE.
Let be the matrix square root of , i.e. .
Define a new regression equation by multiplying both sides by :
Your Turn
Show .
Show the least squares estimates for the new regression equation reduce to:
Can also show .
The estimates: are known as estimates.
In practice, might only be know up to a few parameters that also need to be estimated.
Common cases of GLS
- defines a temporal or spatial correlation structure
- defines a grouping structure
- is diagonal and defines a weighting structure ()
In R
?lm # use weights argument
library(nlme)
?gls # has weights and/or correlation argumentOat yields
Data from an experiment to compare 8 varieties of oats. The growing area was heterogeneous and so was grouped into 5 blocks. Each variety was sown once within each block and the yield in grams per 16ft row was recorded.
library(nlme)
fit_gls <- gls(yield ~ variety, data = oatvar,
correlation = corCompSymm(form = ~ 1 | block))Oat yields in R
intervals(fit_gls)## Approximate 95% confidence intervals
##
## Coefficients:
## lower est. upper
## (Intercept) 291.542999 334.4 377.2570009
## variety2 -4.903898 42.2 89.3038984
## variety3 -18.903898 28.2 75.3038984
## variety4 -94.703898 -47.6 -0.4961016
## variety5 57.896102 105.0 152.1038984
## variety6 -50.903898 -3.8 43.3038984
## variety7 -63.103898 -16.0 31.1038984
## variety8 2.696102 49.8 96.9038984
## attr(,"label")
## [1] "Coefficients:"
##
## Correlation structure:
## lower est. upper
## Rho 0.06596382 0.3959955 0.7493731
## attr(,"label")
## [1] "Correlation structure:"
##
## Residual standard error:
## lower est. upper
## 33.39319 47.04679 66.28298Lack of fit F-tests
should be (if our model is specified correctly) an unbiased estimate of .
A “model free” estimate of is available if there are replicates (multiple observations at combinations of the explanatory values).
If our from our model is much bigger than the “model-free” estimate, we have evidence of .
In practice
Fit a saturated model. Compare saturated model to proposed model with an F-test. .
Saturated: every combination of explanatory variables is allowed its own mean (i.e. every group of replicates is allowed its own mean). A model that includes every explantory as categorical and every possible interaction between variables.
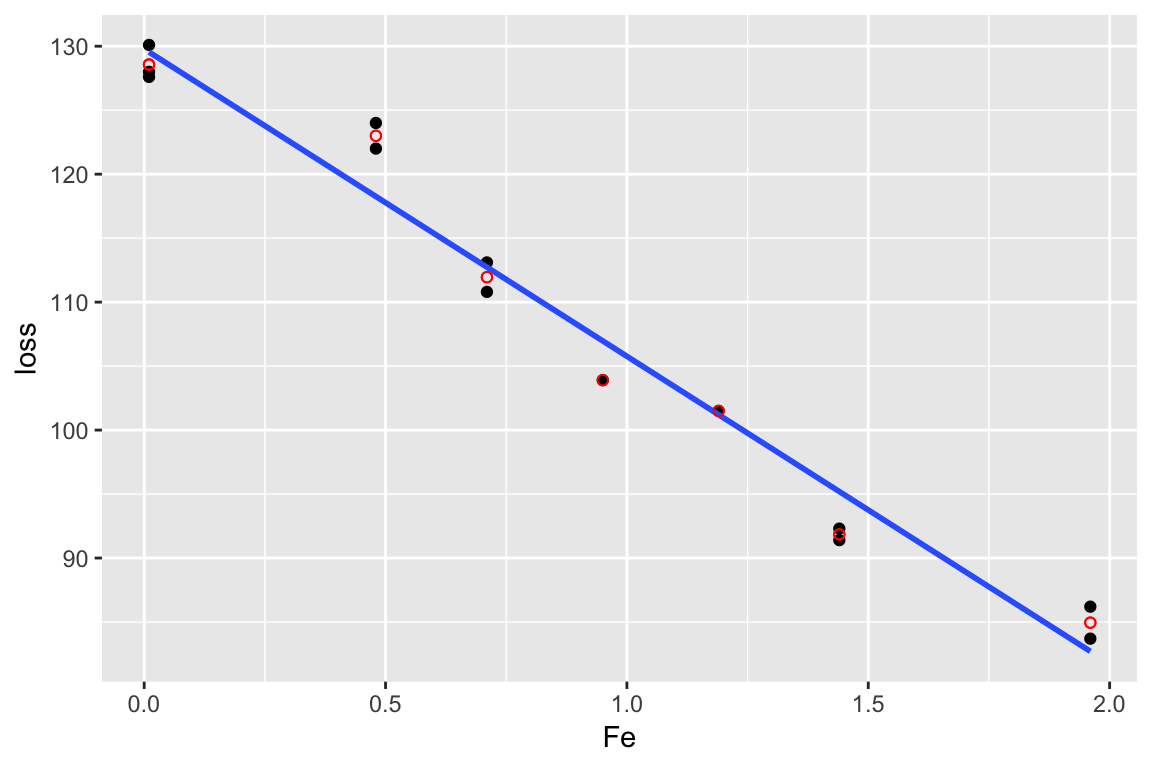
Example
data(corrosion, package = "faraway")
lm_cor <- lm(loss ~ Fe, data = corrosion)
lm_sat <- lm(loss ~ factor(Fe), data = corrosion)
anova(lm_cor, lm_sat)## Analysis of Variance Table
##
## Model 1: loss ~ Fe
## Model 2: loss ~ factor(Fe)
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 11 102.850
## 2 6 11.782 5 91.069 9.2756 0.008623 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1# significant lack of fitRobust regression
Remember to define our least squares estimates we looked for to minimise
In practice, since we are squaring residuals, observations with large residuals carry a lot of weight. For, robust regression, we want to downweight the observations with large residuals.
The idea of M-estimators is to extend this to the general situation where we want to find to minimise where is some function we specify.
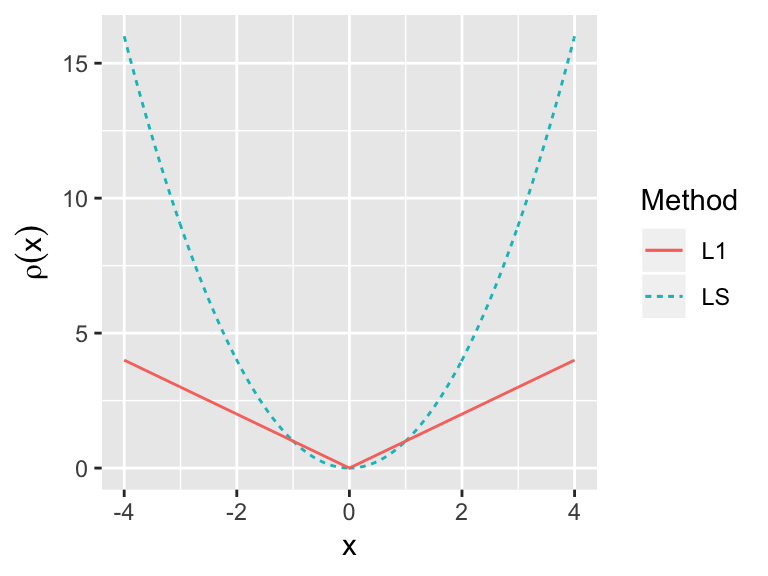
- Least squares:
- Least absolute deviation, L regression:
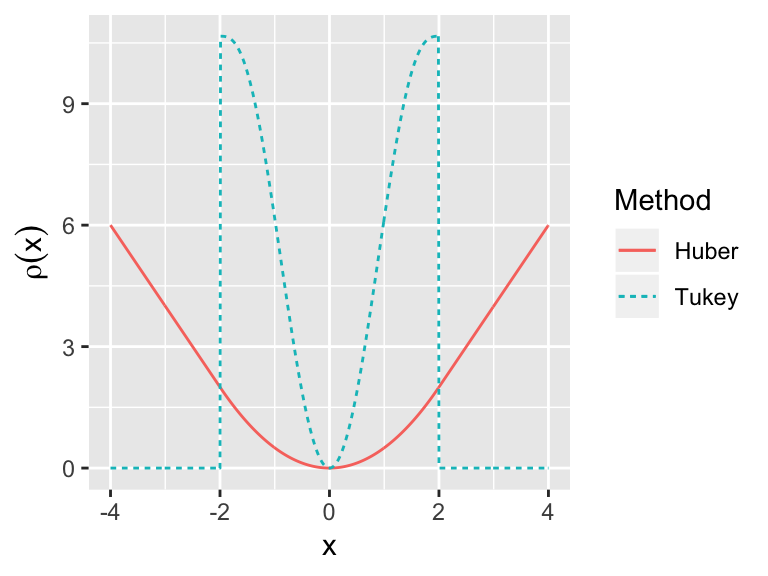
- Huber’s method
- Tukey’s bisquare
The models are usually fit in an iterative process.
Least trimmed squares
Minimise the smallest residuals where is some number smaller than and is the ith smallest residual.
One choice,
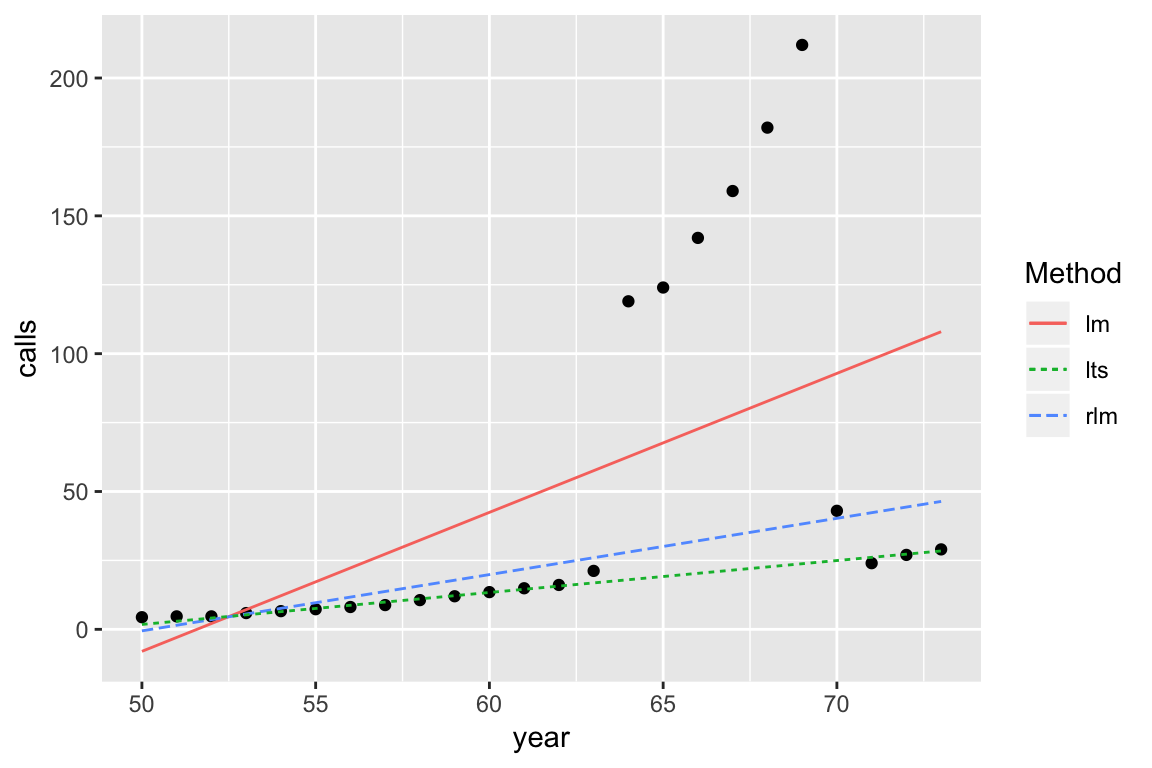
Annual numbers of telephone calls in Belgium